Although the incorporation of artificial intelligence (AI) in antibody research is still in its early stages, research scientists studying AI, bioinformatics, and protein engineering have used AI models to demonstrate its potential to transform the R&D space of the biotech and pharmaceutical industries.
The accessibility of high-throughput experimental data has also enabled a shift in antibody research from being experiment-focused to data-centric. AI models like CNN and LSTM have been used to investigate the sequence, structure, and functionality of antibodies, as well as AI-fueled protein structure modeling platforms like AlphaFold2.1
CNN
A convolutional neural network (CNN) is a deep learning model that is useful for analyzing visual imagery. It utilizes convolutional layers (sliding filters) across the input domain, transforming the input to degrees of filter matching for each filter. A protein structure can be rendered into a compatible format for the convolutional filters, such as a graph, manifold or 3D voxel grid.2
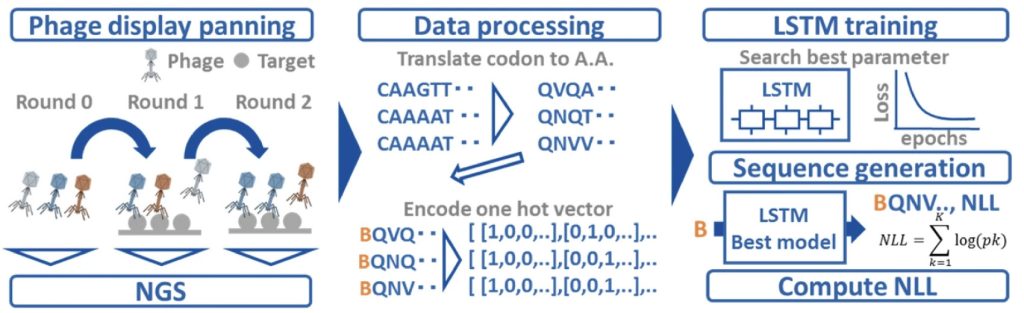
LSTM
A long short-term memory (LSTM) network is another deep learning model that is designed to process and make predictions based on sequences of data. It uses memory cells and gates to capture important information and predict what comes next in the sequence, even when there are long-term dependencies or patterns.3
Mason et al. (2021) used CNNs and LSTMs for antibody lead optimization, successfully predicting antigen specificity from a diverse space of antibody sequences. By deep-sequencing libraries of the existing therapeutic antibody trastuzumab, they generated approximately 1 × 104 variants via site-directed mutagenesis to screen for specificity to human epidermal growth factor receptor 2 (HER2). The sequences were used as input to CNNs and LSTMS which were trained to predict the binding label of the sequences. They used these neural networks to filter a computationally generated set of candidate sequences to HER2-specific predicted binders. By testing for viscosity, clearance, solubility and immunogenicity, the researchers generated thousands of highly optimized lead candidates. Experimental testing revealed 30 out of 30 randomly selected predicted variants retained specificity to the target antigen HER2.4
Therefore, sequence-based methods may be advantageous over traditional structure-based methods, which are more time-consuming and resource-demanding due to the low-throughput screening of full-length antibodies. Meanwhile, sequence-based methods have more data available from developed experimental methods such as next-generation sequencing.
At Biointron, we are dedicated to accelerating your antibody discovery, optimization, and production needs. Our team of experts can provide customized solutions that meet your specific research needs. Contact us to learn more about our services and how we can help accelerate your research and drug development projects.
- Antibody Therapeutics. (2022, February 23). Call for Papers: Artificial Intelligence in antibody discovery, development and beyond. https://academic.oup.com/abt/pages/call-for-papers-artificial-intelligence-in-antibody-discovery-development-and-beyond
- Graves, J., Byerly, J., Priego, E., Makkapati, N., Parish, S. V., Medellin, B., & Berrondo, M. (2020). A Review of Deep Learning Methods for Antibodies. Antibodies, 9(2). https://doi.org/10.3390/antib9020012
- Sepp Hochreiter, Jürgen Schmidhuber; Long Short-Term Memory. Neural Comput 1997; 9 (8): 1735–1780. doi: https://doi.org/10.1162/neco.1997.9.8.1735
- Mason, D. M., Friedensohn, S., Weber, C. R., Jordi, C., Wagner, B., Meng, S. M., Ehling, R. A., Bonati, L., Dahinden, J., Gainza, P., Correia, B. E., & Reddy, S. T. (2021). Optimization of therapeutic antibodies by predicting antigen specificity from antibody sequence via deep learning. Nature Biomedical Engineering, 5(6), 600-612. https://doi.org/10.1038/s41551-021-00699-9