In addition to isotypes and subtypes, antibodies exhibit genetic variation known as allotypes, which are polymorphic epitopes on immunoglobulins. These allotypic differences arise from allelic variations in immunoglobulin genes, causing certain antibody subtypes to differ between individuals or ethnic groups. The presence of these polymorphic forms can influence immune responses, particularly when an individual is exposed to a non-self allotype, potentially triggering an anti-allotype immune reaction.
These immunoglobulin polymorphisms were first identified and referred to as “allotypes” in 1956, following their discovery using rabbit anti-sera produced from animals inoculated with serum from genetically distinct counterparts. Nearly three decades later, monoclonal anti-allotype antibody detectors were developed. Given the immunogenic nature of allotypes, it is not surprising that these alleles influence the interactions between immunoglobulins and their respective anti-isotype detection reagents. Although individual allotypic markers typically involve only one or two amino acid substitutions, these changes can alter protein-protein interactions by modifying the epitope recognized by the detection antibody, hindering binding through steric interference, or inducing conformational changes at the binding site of the detection antibody, even if the epitope is distant from the substituted amino acids. Despite the initial identification of allotypes through serological methods, comprehensive validation of binding by antibody detection reagents to different Ig allotypes is not routinely performed.1
However, not all allotypic variations are immunogenic. Some sequence variations may be present in multiple isotypes or subtypes and are thus classified as isoallotypic variants. Recent studies suggest that allotypic differences in human IgG1, for example, do not pose a significant risk for inducing immunogenicity. This is particularly important in therapeutic contexts, where the use of monoclonal antibodies, such as adalimumab and infliximab, has not shown significant evidence of anti-allotype responses.
G1m Allotypes: Variation in IgG1
Allotypes have been most frequently identified in the gamma 1 (g1), gamma 3 (g3), and alpha 2 (a2) heavy chains, designated as G1m, G3m, and A2m allotypes, respectively. Additionally, allotypic variations are found in the kappa light chain (Km allotypes). Some isoallotypic variants exist in the gamma 2 (g2) and gamma 4 (g4) heavy chains, where the amino acid sequence is shared with other subclasses.
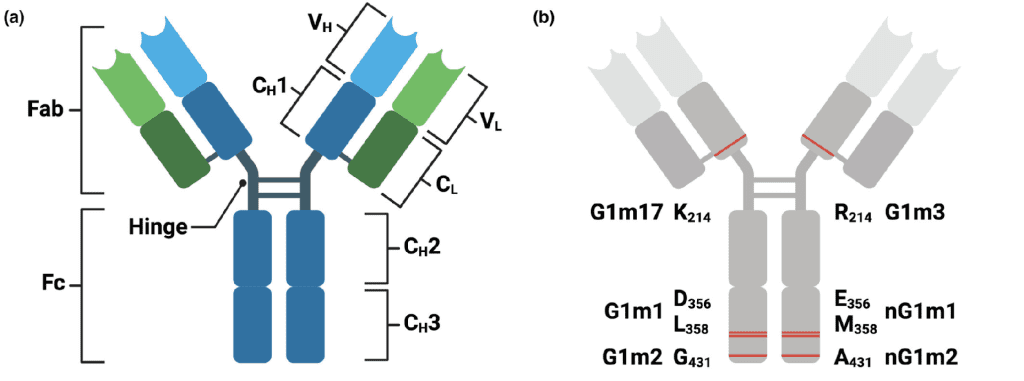
One of the most studied allotypes is G1m, found on the IgG1 heavy chain. These allotypic variants are identified based on specific amino acid changes in the constant region of the antibody. For example, G1m17 (also known as G1m(z)) refers to a lysine (K) at position 214 in the CH1 domain, while G1m3 (G1m(f)) refers to an arginine (R) at the same position. These small changes may seem insignificant, but they provide diversity in the immune response and potential interaction with other proteins or immune cells.
Other notable G1m allotypes include G1m1 and nG1m1, which involve aspartic acid (D) and leucine (L) at positions 356 and 358 in the CH3 domain (EU numbering), as well as G1m2 and nG1m2, involving glycine (G) and alanine (A) at position 431 in the CH3 domain.
Impact of Allotypes on Therapeutic Antibodies
The presence of allotypic variation in therapeutic antibodies raises questions about potential immunogenicity in diverse patient populations. Given that therapeutic monoclonal antibodies are typically derived from human or humanized sources, differences in patient allotypes could theoretically trigger immune responses. However, studies on therapeutic antibodies, such as adalimumab and infliximab, have shown minimal evidence of significant anti-allotype immune responses. This suggests that while allotypes introduce genetic diversity, they do not always translate into clinical challenges for antibody-based therapies.
This lack of substantial anti-allotype response in therapeutic settings may be due to the fact that many allotypic variants do not produce foreign epitopes, or they are isoallotypic and already present across various antibody subclasses. Thus, the risk of inducing immunogenicity due to allotypic variation remains relatively low, providing reassurance for the use of monoclonal antibodies in treating a wide range of conditions, from autoimmune diseases to cancer.
Related: Allotypes
Clinical and Biological Relevance of Allotypes
Allotypes have practical implications in the field of transfusion medicine, transplantation, and antibody-based therapies. Since antibodies with non-self allotypes can induce immune reactions, particularly in individuals with high exposure to non-self proteins, monitoring and understanding allotype variation can be crucial.
For example, in therapeutic antibody production, selecting an antibody variant that matches the allotypic profile of the target population could reduce the potential for immune rejection. Additionally, blood transfusions and organ transplants between individuals with different allotypes may sometimes necessitate special precautions to prevent allotype-related immune complications.
Another biological aspect of allotypes lies in their ability to provide diversity in the immune system’s ability to recognize and neutralize pathogens. Although the differences in the amino acid sequences are small, allotypes may influence how antibodies bind to antigens or Fc receptors, potentially impacting how effectively they initiate immune responses like ADCC (antibody-dependent cellular cytotoxicity) or complement activation.
G3m, A2m, and Km Allotypes
Beyond G1m allotypes, other allotypic variations have been identified in different immunoglobulin subclasses. G3m allotypes are located on the IgG3 subclass, which is notable for its extended hinge region that enhances its flexibility and immune activation potential. A2m allotypes are found in the IgA2 subclass, primarily involved in mucosal immunity. Km allotypes, located on kappa light chains, further contribute to the variability of antibody responses in the population.
Each of these allotypes adds layers of complexity to the immunoglobulin gene pool, influencing not only individual immune responses but also clinical outcomes in immunotherapy, vaccination, and disease susceptibility.
- Purcell, R. A., Aurelia, L. C., Esterbauer, R., Allen, L. F., Bond, K. A., Williamson, D. A., Trevillyan, J. M., Trubiano, J. A., Juno, J. J., Wheatley, A. K., Davenport, M. P., Nguyen, T. H., Kedzierska, K., Kent, S. J., Selva, K. J., & Chung, A. W. (2024). Immunoglobulin G genetic variation can confound assessment of antibody levels via altered binding to detection reagents. Clinical & Translational Immunology, 13(3), e1494. https://doi.org/10.1002/cti2.1494