Understanding the structures and functions is essential to designing an antibody for a specific goal. Research into developing deep learning models which can help to predict these structures is a fascinating, ever-evolving topic. Deep learning is a subset of machine learning based on stacked artificial neural network layers, in which processing is used to extract increasingly higher-level features from affine transformation and non-linear activation function data.1
DeepH3 is a deep learning model that predicts inter-residue distances and orientations from antibody heavy and light chain sequence. These result in distributions which are then converted to geometric representations and used to discriminate between decoy structures and predict new antibody hypervariable complementarity-determining region (CDR) H3 loops de novo.2
These CDR H3 loops are necessary for antigen binding, and they possess high conformational diversity even if there is high sequence similarity.3 Due to this, CDR-H3 modeling has much fewer constraints than the other five of six CDRs (H1, H2, L1, L2, and L3), which means identifying high-quality H3 templates can be a harder job, with DeepH3 models being very valuable.
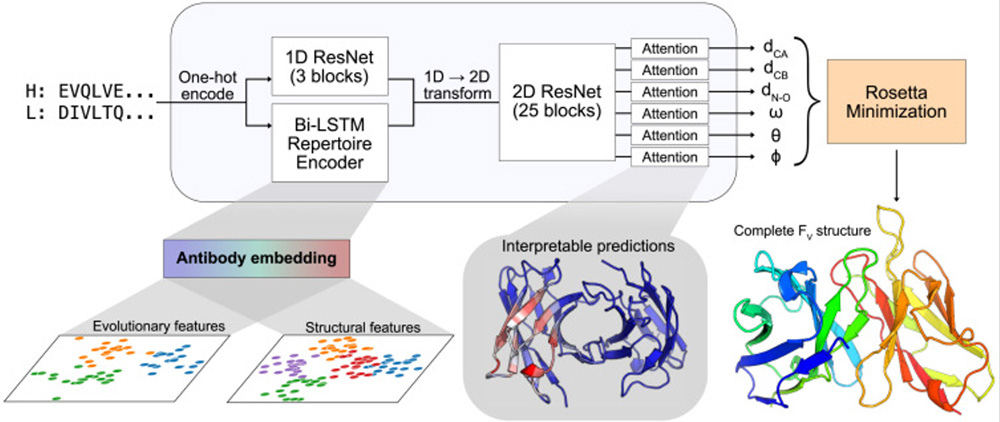
Another example of a deep learning method for antibody structure is DeepAb, which predicts accurate antibody FV structures from sequence. There are two steps to Ruffolo et al.’s (2022) process, with the first using a deep residual convolutional network to predict FV structure. Secondly, they use a quick Rosetta-based protocol to realize the structure from the predictions of the network. This method can also be used to model single-domain (VHH) antibodies.4
At Biointron, we are dedicated to accelerating your antibody discovery, optimization, and production needs. Our team of experts can provide customized solutions that meet your specific research needs. Contact us to learn more about our services and how we can help accelerate your research and drug development projects.
- Gao, W., Mahajan, S. P., Sulam, J., & Gray, J. J. (2020). Deep Learning in Protein Structural Modeling and Design. Patterns, 1(9), 100142. https://doi.org/10.1016/J.PATTER.2020.100142
- Ruffolo, J. A., Guerra, C., Mahajan, S. P., Sulam, J., & Gray, J. J. (2020). Geometric potentials from deep learning improve prediction of CDR H3 loop structures. Bioinformatics, 36(Supplement_1), i268–i275. https://doi.org/10.1093/bioinformatics/btaa457
- Kim, J., McFee, M., Fang, Q., Abdin, O., & Kim, P. M. (2023). Computational and artificial intelligence-based methods for antibody development. Trends in Pharmacological Sciences, 44(3), 175–189. https://doi.org/10.1016/J.TIPS.2022.12.005
- Ruffolo, J. A., Sulam, J., & Gray, J. J. (2022). Antibody structure prediction using interpretable deep learning. Patterns, 3(2), 100406. https://doi.org/10.1016/J.PATTER.2021.100406