Before the road towards clinical trials, the developability properties of therapeutic antibodies need to be evaluated. Antibody molecules are assessed for developability characteristics including aggregation, immunogenicity, pharmacokinetic clearance, viscosity, and half-life span.1 This is important to ensure a stable and efficacious drug before clinical trials.
These properties can be understood through in silico methods, such as homology modelling, docking or interface prediction. During the Lead Identification and Optimization phases these tools can help to researchers to generate three-dimensional models of the antibodies and predict or identify the key residues involved in antigen binding. Machine learning tools can also predict these properties by assessing hydrophobicity, electrostatic charge, and topology pattern interaction of the amino acid sequence.2,3
Computational Tools
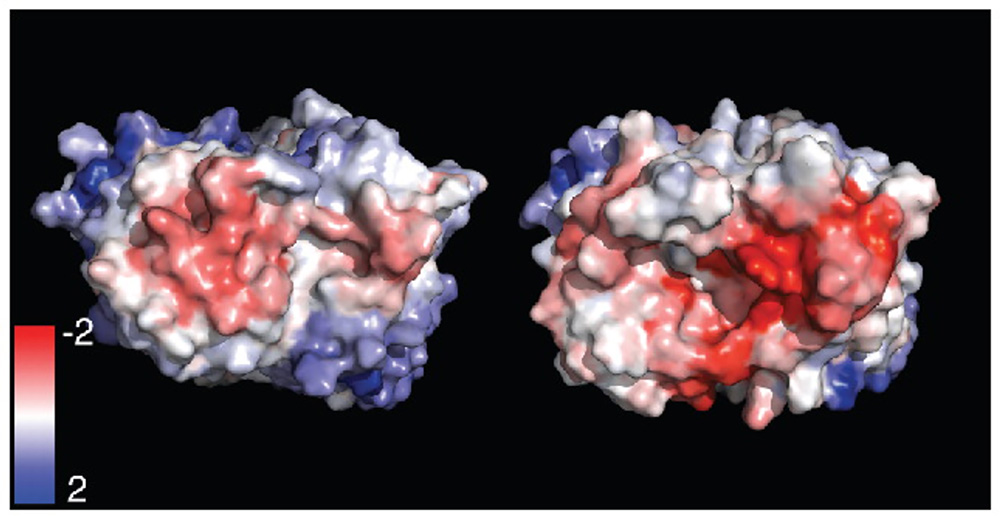
Spatial-Charge-Map (SCM) is a tool developed by Agrawal et al. (2016) that can screen monoclonal antibodies for their viscosities. SCM works by quantifying the electronegative potential patch on the Fv domain of antibodies. High viscosities, commonly found with highly concentrated antibody solutions, poses several problems to develop, manufacture, and administer the potential drug. Since the antibody sequence is a component for viscosity, this SCM tool is extremely helpful for the selection of low-viscosity antibodies.4
The therapeutic antibody profiler (TAP) is an application which compares your antibody variable domain sequence against five factors: total complementarity-determining region (CDR) length, surface hydrophobicity, asymmetry in net heavy- and light-chain surface charges, and positive charge and negative charge in the CDRs. Raybould et al. (2019) published this tool to reveal antibodies that have rare characteristics in clinical-stage mAb therapeutics.5,6
Machine Learning Tools
Jain et al. (2017) used a random forest machine learning approach to estimate the hydrophobic chromatography (HIC) retention time directly from an antibody sequence. Quantifying hydrophobicity of a monoclonal antibody is important to assess downstream risks. The researchers successfully developed predictive models to predict the surface exposure of amino-acid side-chains in the antibody’s variable region.7
At Biointron, we are dedicated to accelerating your antibody discovery, optimization, and production needs. Our team of experts can provide customized solutions that meet your specific research needs. Contact us to learn more about our services and how we can help accelerate your research and drug development projects.
- Raybould, M.I.J., Deane, C.M. (2022). The Therapeutic Antibody Profiler for Computational Developability Assessment. In: Houen, G. (eds) Therapeutic Antibodies. Methods in Molecular Biology, vol 2313. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-1450-1_5
- Kim, J., McFee, M., Fang, Q., Abdin, O., & Kim, P. M. (2023). Computational and artificial intelligence-based methods for antibody development. Trends in Pharmacological Sciences, 44(3), 175–189. https://doi.org/10.1016/J.TIPS.2022.12.005
- Mieczkowski, C., Zhang, X., Lee, D., Nguyen, K., Lv, W., Wang, Y., Zhang, Y., Way, J., & Gries, M. (2023). Blueprint for antibody biologics developability. MAbs, 15(1). https://doi.org/10.1080/19420862.2023.2185924
- Agrawal, N. J., Helk, B., Kumar, S., Mody, N., Sathish, H. A., Samra, H. S., Buck, P. M., Li, L., & Trout, B. L. (2016). Computational tool for the early screening of monoclonal antibodies for their viscosities. MAbs, 8(1), 43-48. https://doi.org/10.1080/19420862.2015.1099773
- J. Raybould, M. I., Marks, C., Krawczyk, K., Taddese, B., Nowak, J., Lewis, A. P., Bujotzek, A., Shi, J., & Deane, C. M. (2019). Five computational developability guidelines for therapeutic antibody profiling. Proceedings of the National Academy of Sciences of the United States of America, 116(10), 4025-4030. https://doi.org/10.1073/pnas.1810576116
- James Dunbar and others, SAbPred: a structure-based antibody prediction server, Nucleic Acids Research, Volume 44, Issue W1, 8 July 2016, Pages W474–W478, https://doi.org/10.1093/nar/gkw361
- Jain, T., Boland, T., Lilov, A., Burnina, I., Brown, M., Xu, Y., & Vásquez, M. (2017). Prediction of delayed retention of antibodies in hydrophobic interaction chromatography from sequence using machine learning. Bioinformatics, 33(23), 3758-3766. https://doi.org/10.1093/bioinformatics/btx519