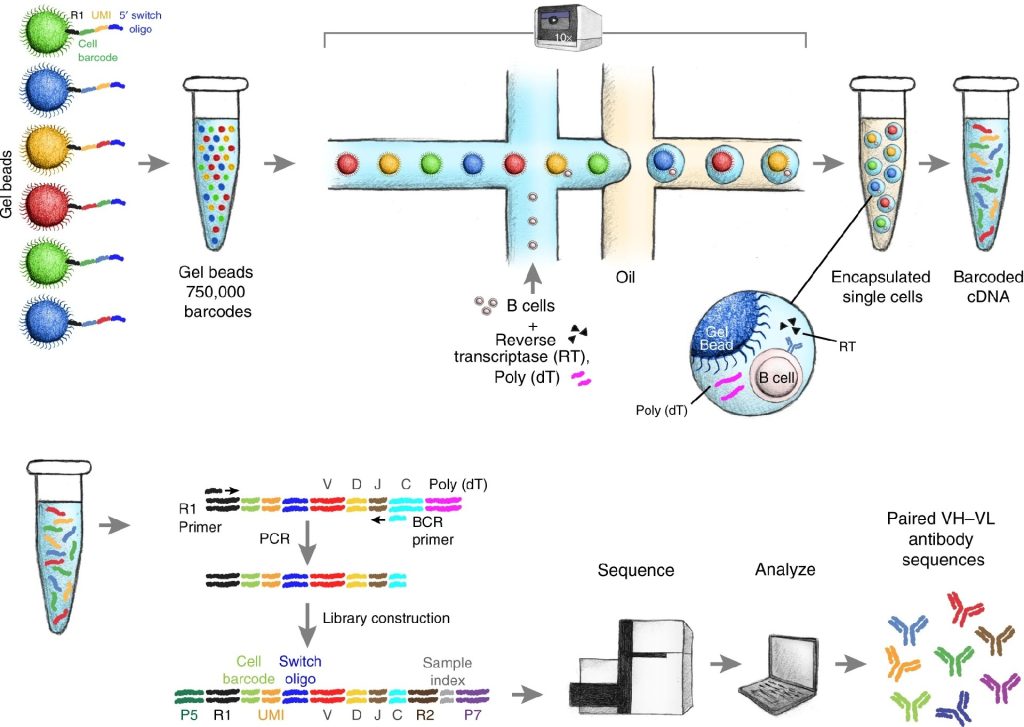
Single-cell antibody sequencing is a technique for analyzing the genome of individual antibody-producing cells, particularly B cells. This method provides detailed insights into the diversity of the immune response by isolating and sequencing the genetic material of single cells, allowing researchers to overcome the limitations of bulk sequencing. With the ability to sequence individual cells, single-cell antibody sequencing reveals the heterogeneity in antibody repertoires, offering a picture of how B cells generate antibodies in response to different antigens.
How Single-Cell Antibody Sequencing Works
In single-cell antibody sequencing, researchers begin by isolating individual antibody-producing cells, such as B cells, from a heterogeneous cell population. Once isolated, the genome of each single cell is amplified to create sufficient material for sequencing. This amplification process is essential because each B cell produces a unique antibody through a combination of heavy and light chain genes. Amplifying and sequencing these genes provides the necessary data to map the full-length variable regions that determine the antibody’s specificity and affinity for its target antigen.
For example, high-throughput single-cell B-cell receptor sequencing (scBCR-seq) has been used to obtain accurately paired full-length variable regions in a massively parallel fashion.1 This overcomes limitations of diversity or heterogeneity that is hidden by isolating and sequencing the genetic material from individual B cells.
By sequencing the entire variable region, researchers can investigate the specific genetic mutations and somatic hypermutations that have occurred during the immune response. These mutations are key to the process of affinity maturation, where B cells fine-tune their antibodies to bind more effectively to a target.
Related: What is Affinity Maturation?
Overcoming the Challenges of Bulk Sequencing
Bulk sequencing methods, which involve sequencing the genetic material from a large population of B cells at once, can mask the true diversity of the immune repertoire. This is because bulk sequencing provides an averaged picture of the antibody repertoire, without distinguishing the individual contributions of different B cells. In contrast, single-cell antibody sequencing preserves the distinct identities of individual B cells, revealing the heterogeneity that is crucial for understanding the full scope of the immune response.
For example, bulk sequencing often struggles to correctly pair heavy and light chains, which can lead to incomplete or misleading representations of the antibody structure. Since an antibody’s specificity relies on the precise pairing of these chains, inaccurate pairing can hinder efforts to understand or engineer new antibodies for therapeutic use. Single-cell sequencing solves this issue by ensuring that the heavy and light chains sequenced come from the same individual B cell, offering a more reliable picture of antibody function.
In addition to providing accurate chain pairing, single-cell antibody sequencing also allows researchers to capture rare or low-frequency antibodies that might be missed in bulk sequencing. These rare antibodies could be particularly valuable for therapeutic development, as they might target unique or previously unrecognized epitopes.
- Goldstein, L. D., Chen, Y., Wu, J., Chaudhuri, S., Hsiao, Y., Schneider, K., Hoi, K. H., Lin, Z., Guerrero, S., Jaiswal, B. S., Stinson, J., Antony, A., Pahuja, K. B., Seshasayee, D., Modrusan, Z., Hötzel, I., & Seshagiri, S. (2019). Massively parallel single-cell B-cell receptor sequencing enables rapid discovery of diverse antigen-reactive antibodies. Communications Biology, 2(1), 1-10. https://doi.org/10.1038/s42003-019-0551-y